Basic time series analysis
In this chapter, we learn some basic time series analysis techniques and how to read real world data from a single file and from a batch of files.
Generating a test time series
Before we start to analyze time series, we first have a look on how they are generated. We now construct a very simple time series from scratch with Python.
Create a script including the following commands:
# Create some dummy data:
import numpy as np
import matplotlib.pyplot as plt
# imports most relevant Matplotlib commands
# create x-values:
dx = 0.1
x_values = np.arange(0, 2*np.pi, dx)
# define parameters:
f1 = 2 # Frequency 1 in Hz
f2 = 10 # Frequency 2 in Hz
A1 = 6 # Amplitude 1
A2 = 2 # Amplitude 2
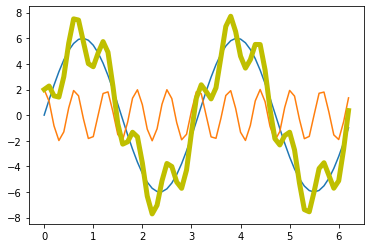
A_sin = A1 * np.sin(f1 * x_values)
A_cos = A2 * np.cos(f2 * x_values)
A_signal = A_sin + A_cos
# plots:
fig=plt.figure(1)
plt.plot(x_values, A_sin)
plt.plot(x_values, A_cos)
plt.plot(x_values, A_signal, lw=5, c="y")
plt.show()
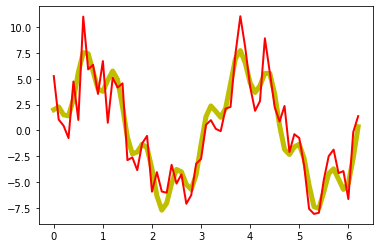
Now, we add some noise to the signal:
# add some noise:
np.random.seed(1)
A_Noise = 2
Noise = np.random.randn(len(x_values)) * A_Noise
A_signal_noisy = A_signal + Noise
fig=plt.figure(1)
plt.plot(x_values, A_signal, lw=5, c="y")
plt.plot(x_values, A_signal_noisy, lw=2, c="r")
plt.show()
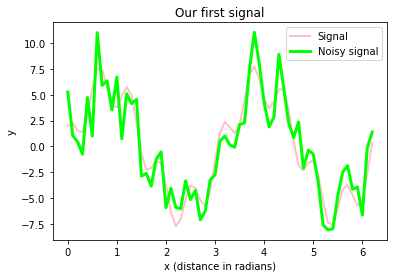
Exercise 1
Basing the solution of Exercise 2 from the Data Visualization with Matplotlib chapter in the Python Basics course, copy and modify the appropriate plot commands, in order to plot
A_signal(add the following properties to the plot command:label="Signal", lw=2, c="pink")A_signal_noisy(add the following properties to the plot command:label="Noisy signal", lw=3, c="lime")
in a single plot window. Don’t forget to annotate your plot with appropriate x- and y-labels, a title, and a legend. Also, save your plot as a PDF.
# Your solution 1 here:
Toggle solution
# Solution 1:
# Plotting:
fig = plt.figure(1)
fig.clf()
# the main plot commands:
plt.plot(x_values, A_signal,
label="Signal", lw=2, c="pink")
plt.plot(x_values, A_signal_noisy,
label="Noisy signal", lw=3, c="lime")
# axis labels and title:
plt.xlabel("x (distance in radians)")
plt.ylabel("y")
plt.title("Our first signal")
# shows a legend:
plt.legend(loc="best")
plt.show() # finalizes the plot
fig.savefig("my_plot2.pdf", dpi=120)The figure above includes
- our signal, which is a super-position of actually two prime signals (a sine and a cosine wave with different frequencies and amplitudes)
- our signal with some random (“background”) noise.
Signal filtering
We will now try to filter our signal in order to
- recover the first and the second prime signal, respectively.
- apply the same filtering operations to the noisy signal.
Let’s first apply a so-called butter-worth filter to recover the (lower frequency) sine signal (i.e., we perform low-pass filtering). For this, we use the butter function from the scipy-filter package:
from scipy import signal # put this on top of
# your script
# set filter parameters:
fs = x_values.shape[0] # Sampling frequency
fc = 5 # Cut-off frequency of the filter
w = fc / (fs / 2) # Normalize the frequency
filter_order = 10
# apply the filter function:
b, a = signal.butter(filter_order, w, 'low')
# filtering our signal
A_signal_clean = signal.filtfilt(b, a, A_signal)
plt.plot(x_values, A_signal, label='Signal', c="k")
plt.plot(x_values, A_signal_clean, label='filtered',
c = "lime", lw=4)
plt.plot(x_values, A_sin, label='Sine', c="blue")
plt.plot(x_values, A_cos, label='Cosine', c="red")
plt.legend(loc="best")
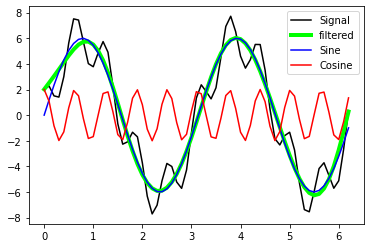
With the defined filter settings we are able to recover the sine-signal from A_signal.
In order to create a corresponding high-pass filter to also recover the high-frequency cosine signal, we would have to repeat all filter relevant commands from above. That’s actually not the way how we should do. Instead, it would be convenient if we would have our own filter-function, so that we don’t have to repeat all these complex steps.
Exercise 2: Construct a filter function
- Recap the Function definition chapter from our Python Basics course.
- Write a function that performs the low-pass filtering from above. Define the function in this way that the following arguments must be passed as input parameters:
sampling_freqfor the sampling frequency,cutoff_freqfor the cut-off frequency of the filter,filter_orderfor the filter order, andinput_signalfor the input signal, that has to be processedkindfor the type of the filter to be applied (eitherloworhigh)
- Apply your newly created function to
A_signal, once for low-pass filtering the signal, and once for high-pass filtering it, and add corresponding plotting commands to plot- the unimpaired signal
A_signal, - the high-pass filtered signal (e.g.,
A_clean1), - the low-pass filtered signal (e.g.,
A_clean2), and - a cosine and a sine wave for comparison.
- the unimpaired signal
- Apply your filter to the noisy signal
A_signal_noisy, again both for low- and high-pass filtering the signal. In a new plot window, plot,- the noisy signal
A_signal_noisy, - the high-pass filtered signal (e.g.,
A_clean1_noisy), - the low-pass filtered signal (e.g.,
A_clean2_noisy), and - a cosine and a sine wave for comparison. What do you notice?
- the noisy signal
# Your solution 2.2 here:
Toggle solution
# Solution 2.2:
def my_filter(sampling_freq, cutoff_freq,
filter_order, input_signal, kind):
""" Function for optionally high- or low-pass
filter a given input signal
sampling_freq: Sampling frequency
cutoff_freq: Cut-off frequency of the filter
filter_order: Order of the filter
input_signal: Signal to filter
kind: "high" or "low"
"""
fc = cutoff_freq
fs = sampling_freq
w = fc / (fs / 2) # Normalize the frequency
b, a = signal.butter(filter_order, w, kind)
# filtering the signal
filtered = signal.filtfilt(b, a, input_signal)
return filtered# Your solution 2.3 here:
Toggle solution
# Solution 2.3:
# Apply the filter function:
A_clean1=my_filter(sampling_freq = x_values.shape[0],
cutoff_freq = 5, filter_order = 10,
input_signal = A_signal, kind = "high")
A_clean2=my_filter(sampling_freq = x_values.shape[0],
cutoff_freq = 5, filter_order = 10,
input_signal = A_signal, kind = "low")
# Plotting:
fig = plt.figure(1)
fig.clf()
# the main plot commands:
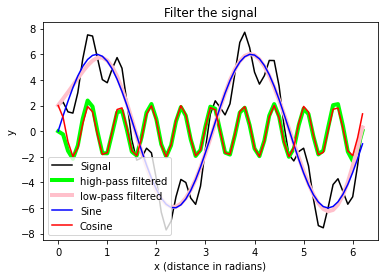
plt.plot(x_values, A_signal, label='Signal', c="k")
plt.plot(x_values, A_clean1, label='high-pass filtered',
c = "lime", lw=4)
plt.plot(x_values, A_clean2, label='low-pass filtered',
c = "pink", lw=4)
plt.plot(x_values, A_sin, label='Sine', c="blue")
plt.plot(x_values, A_cos, label='Cosine', c="red")
plt.legend(loc="best")
# axis labels and title:
plt.xlabel("x (distance in radians)")
plt.ylabel("y")
plt.title("Filter the signal")
# shows a legend:
plt.legend(loc="best")
plt.show() # finalizes the plot
fig.savefig("my_plot3.pdf", dpi=120)# Your solution 2.4 here:
Toggle solution
# Solution 2.4:
A_clean1_noisy=my_filter(sampling_freq = x_values.shape[0],
cutoff_freq = 5, filter_order = 10,
input_signal = A_signal_noisy,
kind = "high")
A_clean2_noisy=my_filter(sampling_freq = x_values.shape[0],
cutoff_freq = 5, filter_order = 10,
input_signal = A_signal_noisy,
kind = "low")
# Plotting:
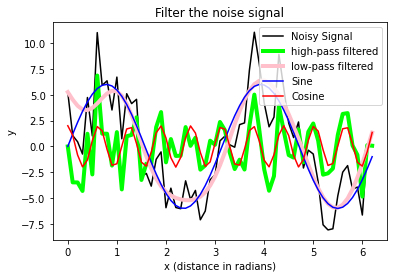
fig = plt.figure(1)
fig.clf()
# the main plot commands:
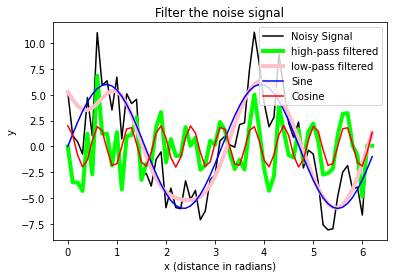
plt.plot(x_values, A_signal_noisy,
label='Noisy Signal', c="k")
plt.plot(x_values, A_clean1_noisy, label='high-pass filtered',
c = "lime", lw=4)
plt.plot(x_values, A_clean2_noisy, label='low-pass filtered',
c = "pink", lw=4)
plt.plot(x_values, A_sin, label='Sine', c="blue")
plt.plot(x_values, A_cos, label='Cosine', c="red")
plt.legend(loc="best")
# axis labels and title:
plt.xlabel("x (distance in radians)")
plt.ylabel("y")
plt.title("Filter the noise signal")
# shows a legend:
plt.legend(loc="best")
plt.show() # finalizes the plot
fig.savefig("my_plot4.pdf", dpi=120)Advice: Matplotlib commands can grow big very quickly. It’s therefore often useful to plug these commands into a function definition in order to keep your script readable and especially when you need to repeat your plot command(s) several times within your script.
Exercise 3: Reading real world data
- Download the data folder “Data/Pandas_3” from the GitHub repository ꜛ (if not already done).
- Write a script, the reads the file “mouse_1_odorA_3_results.xlsx” into a Pandas DataFrame.
-
Iterate over the keys of the DataFrame (here called
df) viafor key in df.keys(): plt.plot(time, df[key])and plot each column of the read Excel file data into one figure. Set an appropriate title and x- and y-labels. Use the sample frequency
sampling_rate = 30.1(unit: [1/s]) to construct the correspondingtimearray:time = np.arange(df.shape[0]) / sampling_rate - Visit the documentation website ꜛ of the
scipy.signal.medfiltand read how to apply a 1D median filter to a given data array. Apply this filter to each column of the data and plot the filtered results into a new figure.
# Your solution 3.2-3.3 here:
Toggle solution
# Solution 3.2-3.3:
import pandas as pd
import os
import numpy as np
import matplotlib.pyplot as plt
from scipy import signal
# %% DEFINE PATHS
"""Define file paths:"""
file_path = "Data/Pandas_2/"
file_name = "ROI_table.xlsx"
file_1 = os.path.join(file_path, file_name)
# %% DATA READING
""" Read the Excel files with Pandas into a Pandas Dataframe:"""
#df = pd.read_excel(file_1, engine='openpyxl')
sheet_to_read = "Tabelle1"
df = pd.read_excel(file_1, index_col=0, sheet_name=sheet_to_read)
sampling_rate = 30.1
time = np.arange(df.shape[0])/sampling_rate
# %% SOME PLOTS
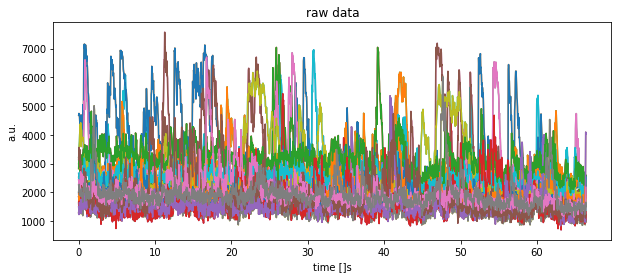
fig=plt.figure(1,figsize=(10,4))
plt.clf()
for key in df.keys():
plt.plot(time, df[key])
plt.title("raw data")
plt.xlabel("time []s")
plt.ylabel("a.u.")
plt.show()
plt.savefig(file_1 + "raw.pdf")# Your solution 3.4 here:
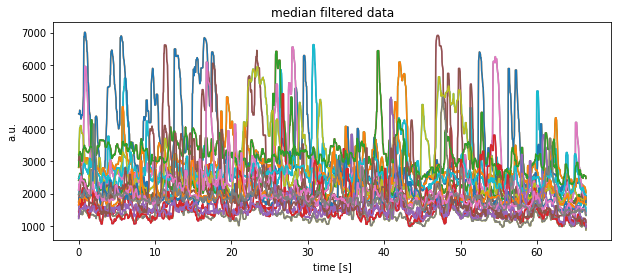
Toggle solution
# Solution 3.4:
fig = plt.figure(2, figsize=(10, 4))
plt.clf()
for key in df.keys():
plt.plot(time, signal.medfilt(df[key], kernel_size=9))
plt.title("median filtered data")
plt.xlabel("time [s]")
plt.ylabel("a.u.")
plt.show()
plt.savefig(file_1 + "median filtered.pdf")df.shape
(2000, 38)
# Alternative solution:
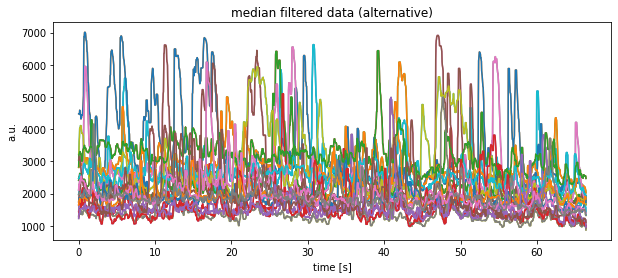
Toggle alternative solution
# Pre-calculate (and save) median filtered data:
df_medianfiltered = np.zeros(df.shape) #This is not a DataFrame
# any more, but a NUMPY
# arrays
for column, key in enumerate(df.keys()):
#print(f"column: {column}, key: {key}")
df_medianfiltered[:, column] = \
signal.medfilt(df[key], kernel_size=9)
fig = plt.figure(2, figsize=(10, 4))
plt.clf()
for column, key in enumerate(df.keys()):
#df_medianfiltered[:, column] = \
# signal.medfilt(df[key], kernel_size=9)
plt.plot(time, df_medianfiltered[:, column])
plt.title("median filtered data (alternative)")
plt.xlabel("time [s]")
plt.ylabel("a.u.")
plt.show()
plt.savefig(file_1 + "median filtered (alt).pdf")Example for exporting a Pandas DataFrame into an Excel file:
# Example for exporting our processed data into a new Excel file:
df_medianfiltered_out_df = pd.DataFrame(data=df_medianfiltered)
df_medianfiltered_out_df.to_excel("data_medianfiltered.xlsx")
"""Note, for initial call of '.to_excel' you need to install
the package/module 'xlwt' """
df_medianfiltered_out_df
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1979.681 | 4476.209 | 2814.321 | 2416.224 | 1648.606 | 1449.719 | 1744.611 | 2119.442 | 1305.099 | 3221.120 | ... | 3221.120 | 2468.984 | 1750.352 | 1649.530 | 2833.220 | 1282.657 | 1231.136 | 3042.830 | 2108.877 | 1966.602 |
| 1 | 2007.620 | 4476.209 | 2838.231 | 2425.859 | 1648.606 | 1606.934 | 1886.981 | 2251.333 | 1305.119 | 3336.423 | ... | 3336.423 | 2477.631 | 1854.227 | 1785.687 | 3217.644 | 1435.793 | 1374.950 | 3142.866 | 2266.636 | 1966.602 |
| 2 | 2079.598 | 4476.209 | 2866.485 | 2520.453 | 1648.606 | 1606.934 | 1938.510 | 2251.333 | 1340.275 | 3628.556 | ... | 3628.556 | 2508.199 | 1875.356 | 1785.687 | 3231.953 | 1457.536 | 1374.950 | 3142.866 | 2266.829 | 2029.088 |
| 3 | 2079.598 | 4476.209 | 2946.522 | 2522.406 | 1737.614 | 1618.653 | 2003.899 | 2321.433 | 1357.278 | 3760.244 | ... | 3760.244 | 2516.503 | 1875.356 | 1807.257 | 3269.945 | 1500.843 | 1425.907 | 3142.866 | 2283.250 | 2029.088 |
| 4 | 2079.598 | 4589.748 | 2948.629 | 2625.568 | 1737.614 | 1618.653 | 2086.211 | 2424.808 | 1419.689 | 4031.759 | ... | 4031.759 | 2575.812 | 1875.356 | 1852.093 | 3296.585 | 1500.843 | 1500.500 | 3142.866 | 2334.456 | 2029.088 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1995 | 1659.978 | 1988.962 | 2204.836 | 1856.755 | 1084.831 | 1065.796 | 1785.423 | 1737.096 | 982.970 | 1787.830 | ... | 1787.830 | 2002.620 | 1782.898 | 1908.837 | 2551.720 | 1140.593 | 1198.836 | 1228.652 | 1450.763 | 1504.574 |
| 1996 | 1623.542 | 1988.962 | 2187.248 | 1768.047 | 1008.979 | 1058.442 | 1749.308 | 1737.096 | 982.970 | 1787.830 | ... | 1787.830 | 2002.620 | 1760.417 | 1908.837 | 2551.720 | 1022.057 | 1198.836 | 1207.696 | 1450.763 | 1504.574 |
| 1997 | 1585.314 | 1981.024 | 2127.415 | 1768.047 | 993.758 | 1018.179 | 1749.308 | 1737.096 | 970.974 | 1697.901 | ... | 1697.901 | 1934.899 | 1760.417 | 1908.837 | 2551.720 | 1018.071 | 1198.836 | 1193.161 | 1421.070 | 1357.963 |
| 1998 | 1581.983 | 1874.429 | 2110.344 | 1757.333 | 993.106 | 1018.179 | 1660.389 | 1737.096 | 957.219 | 1690.157 | ... | 1690.157 | 1934.899 | 1760.417 | 1908.837 | 2546.767 | 1010.943 | 1084.129 | 1185.286 | 1421.070 | 1323.653 |
| 1999 | 1333.852 | 1786.643 | 2025.873 | 1757.333 | 936.424 | 1002.555 | 1660.389 | 1737.096 | 874.662 | 1658.114 | ... | 1658.114 | 1934.899 | 1733.292 | 1836.423 | 2472.458 | 1010.943 | 962.400 | 1105.598 | 1385.715 | 1323.653 |
2000 rows × 38 columns
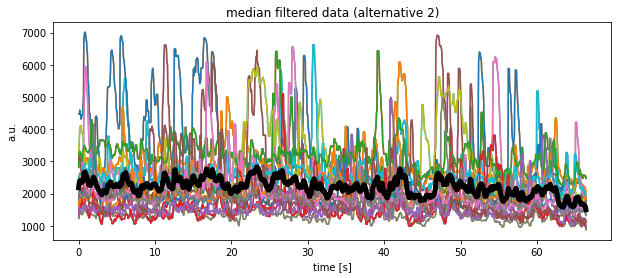
Grand average
# for the sake of curiosity, let's plot the Grand Average:
fig = plt.figure(2, figsize=(10, 4))
plt.clf()
for column, key in enumerate(df.keys()):
plt.plot(time, df_medianfiltered[:, column])
plt.plot(time, df_medianfiltered.mean(axis=1), 'k', lw=5)
plt.title("median filtered data (alternative 2)")
plt.xlabel("time [s]")
plt.ylabel("a.u.")
plt.show()
plt.savefig(file_1 + "median filtered (alt 2).pdf")
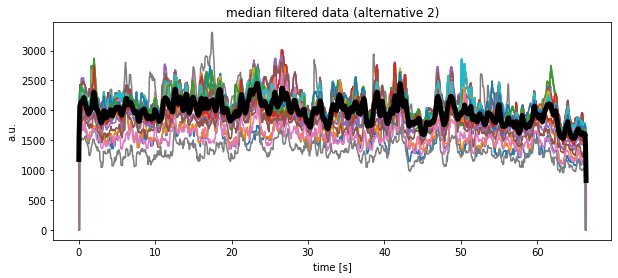
Another alternative solution would look like as follows, but at the moment I do not understand, why the medians look so different compared to the previous calculations?!
# Another alternative solution to 3.4:
Toggle another alternative solution
# Solution 3.4 (Alternative 2):
# Pre-calculate (and save) median filtered data:
df_medianfiltered_3 = signal.medfilt(df, kernel_size=9)
#This is not a DataFrame any more, but a NUMPY array
fig = plt.figure(2, figsize=(10, 4))
plt.clf()
for column in range(df_medianfiltered_3.shape[1]):
plt.plot(time, df_medianfiltered_3[:, column])
plt.plot(time, df_medianfiltered_3.mean(axis=1), 'k', lw=5)
plt.title("median filtered data (alternative 2)")
plt.xlabel("time [s]")
plt.ylabel("a.u.")
plt.show()
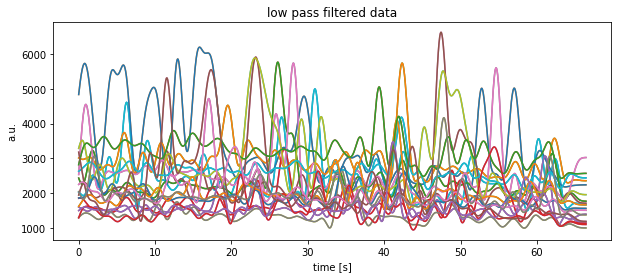
plt.savefig(file_1 + "median filtered (alt 2).pdf")Exercise 4: Apply your filter function to real world data (Exercise 3 continued)
- Apply the
my_filterfunction, that we have written above, to the data read in Exercise 3 and plot, again, the results into a new figure. - Save each of your three figures (those from Exercise 3 including) into a separate PDF file.
# Your solution 4 here:
Toggle solution
# Solution 4:
# recap our previous designed filter-function:
def my_filter(sampling_freq, cutoff_freq,
filter_order, input_signal, kind):
""" Function for optionally high- or low-pass
filter a given input signal
sampling_freq: Sampling frequency
cutoff_freq: Cut-off frequency of the filter
filter_order: Order of the filter
input_signal: Signal to filter
kind: "high" or "low"
"""
fc = cutoff_freq
fs = sampling_freq
w = fc / (fs / 2) # Normalize the frequency
b, a = signal.butter(filter_order, w, kind)
# filtering the signal
filtered = signal.filtfilt(b, a, input_signal)
return filtered
fig = plt.figure(2, figsize=(10, 4))
plt.clf()
for key in df.keys():
current_signal = my_filter(sampling_freq = sampling_rate,
cutoff_freq = 0.5,
filter_order = 10,
input_signal = df[key],
kind = "low")
plt.plot(time, current_signal)
plt.title("low pass filtered data")
plt.xlabel("time [s]")
plt.ylabel("a.u.")
plt.show()
plt.savefig(file_1 + "low-pass filtered.pdf")Exercise 5: Batch file processing
Now, and instead of reading each Excel manually, use the following command to scan for all Excel files in the downloaded folder:
file_name = [f for f in os.listdir(file_path) if f.endswith('.xlsx')]
Copy your solutions from above into a new script. Change the new script in that way, that it iterates over all scanned file names in file_name, starting from the Pandas reading part. Play a bit around, to see, what is contained in file_name.
Hint: The automatized reading of the filenames now requires an iteration of your pipeline within a for-loop.
# Your solution 5 here:
['ROI_table.xlsx', 'ROI_table_1.xlsx', 'ROI_table_5.xlsx', 'ROI_table_4.xlsx', 'ROI_table_3.xlsx', 'ROI_table_2.xlsx']
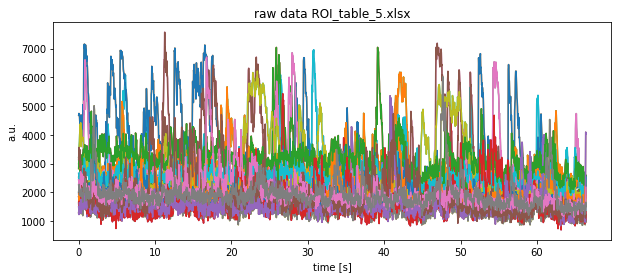
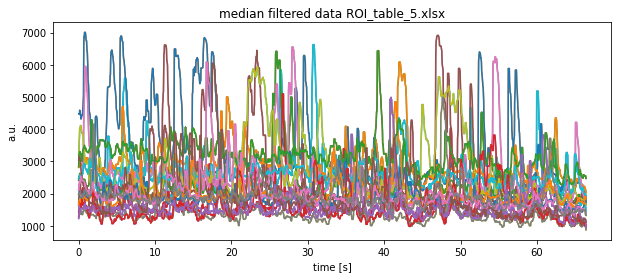
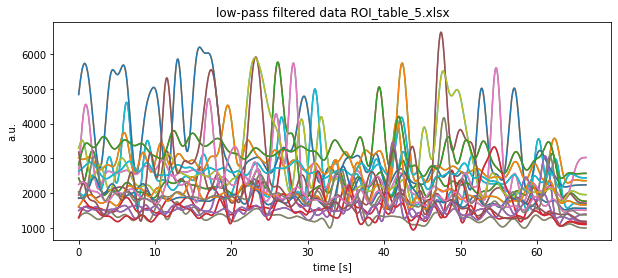
Note that we show the result of only one Excel file here.
Toggle solution
# Solution 5 (full script):
import pandas as pd
import os
import numpy as np
import matplotlib.pyplot as plt
from scipy import signal
# %% DEFINE PATHS
"""Define file paths:"""
file_path = "Data/Pandas_2/"
file_name = [f for f in os.listdir(file_path) if f.endswith('.xlsx')]
sheet_to_read = "Tabelle1"
print(file_name)
# %% SOME FUNCTIONS
def my_filter(sampling_freq, cutoff_freq,
filter_order, input_signal, kind):
""" Function for optionally high- or low-pass
filter a given input signal
sampling_freq: Sampling frequency
cutoff_freq: Cut-off frequency of the filter
filter_order: Order of the filter
input_signal: Signal to filter
kind: "high" or "low"
"""
fc = cutoff_freq
fs = sampling_freq
w = fc / (fs / 2) # Normalize the frequency
b, a = signal.butter(filter_order, w, kind)
# filtering the signal
filtered = signal.filtfilt(b, a, input_signal)
return filtered
# %% ITERATIVELY DATA READING
""" Read the Excel files with Pandas into a Pandas Dataframe:"""
sampling_rate = 30.1 # in Hertz [1/s]
#for file in file_name: # uncomment this line for a full run
for file in [file_name[2]]:
# read the current Excel file:
current_file = os.path.join(file_path, file)
df = pd.read_excel(current_file, index_col=0,
sheet_name=sheet_to_read)
#df = pd.read_excel(current_file, engine='openpyxl')
# calculate the current time-array:
time = np.arange(df.shape[0]) / sampling_rate
# plots:
fig = plt.figure(1, figsize=(10, 4))
plt.clf()
for key in df.keys():
plt.plot(time, df[key])
plt.title(f"raw data {file}")
plt.xlabel("time [s]")
plt.ylabel("a.u.")
plt.show()
plt.savefig(file_path + file + "raw.pdf")
fig = plt.figure(2, figsize=(10, 4))
plt.clf()
for key in df.keys():
plt.plot(time, signal.medfilt(df[key], kernel_size=9))
plt.title(f"median filtered data {file}")
plt.xlabel("time [s]")
plt.ylabel("a.u.")
plt.show()
plt.savefig(file_path + file + "median filtered.pdf")
fig = plt.figure(3, figsize=(10, 4))
plt.clf()
for key in df.keys():
current_signal = my_filter(sampling_freq=sampling_rate,
cutoff_freq=0.5, filter_order=10,
input_signal=df[key], kind="low")
plt.plot(time, current_signal)
plt.title(f"low-pass filtered data {file}")
plt.xlabel("time [s]")
plt.ylabel("a.u.")
plt.show()
plt.savefig(file_path+file+"low pass filtered.pdf")